For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Glutamate Decarboxylase - Manual
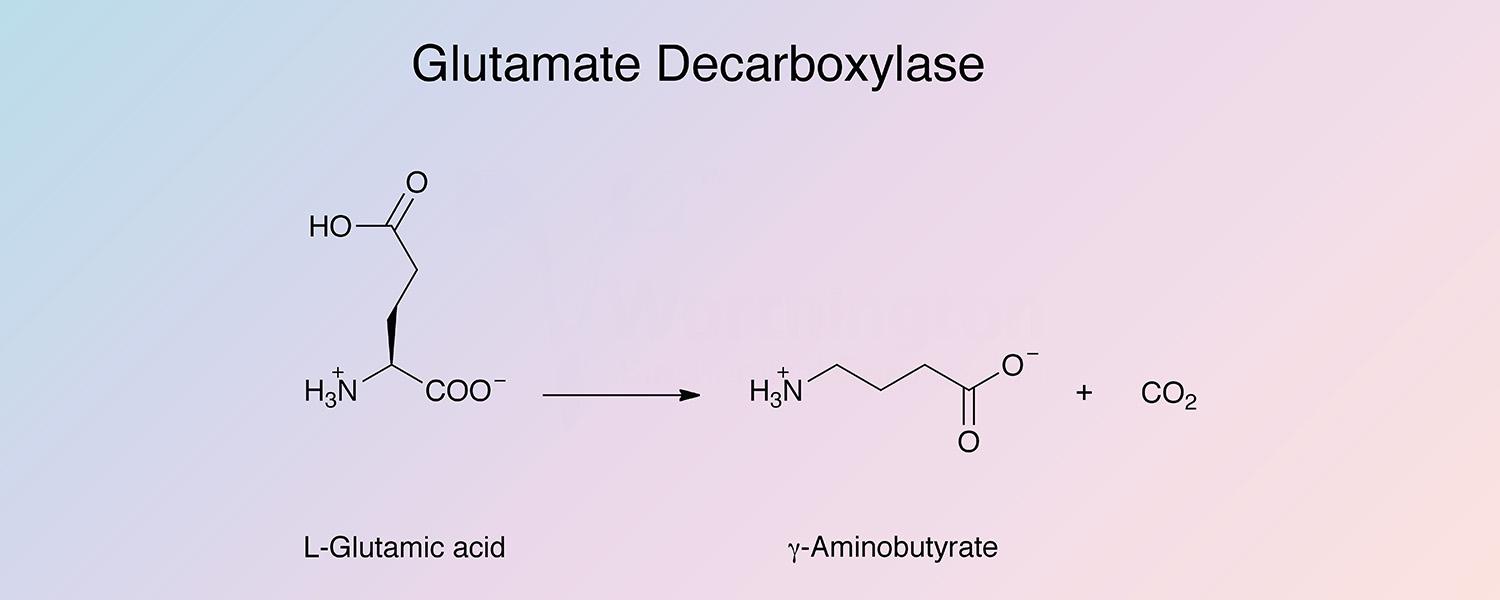
Glutamate decarboxylase (GAD) is a pyridoxal enzyme that catalyzes the removal of the carboxyl group of L-glutamic acid adjacent to the α-amino group to produce γ-aminobutyric acid and carbon dioxide:
![]()
E. coli GAD maintains cellular pH under acidic conditions in the intestines, allowing E. coli to survive gastric acidity and volatile fatty acids produced during fermentation in the intestines. As a result, E. coli can cause disease even when a small amount of cells are ingested. In humans and higher mammals, the enzyme fulfills important physiological functions in the nervous system by balancing excitation and inhibition of neurons. It also acts as a autoantigen in insulin-dependent diabetes (Dutyshev et al. 2005). The following information applies to GAD of E. coli.
Investigations in the early 1900s showed the formation of amines from amino acids by mixing cultures of microorganisms, without knowing the processes, organisms, or enzymes involved (Ellinger 1900, Abderhalden et al. 1913, and Ackermann 1910, 1911). Later work was done using pure strains of identified organisms from the intestine, and the amino acids formed by each organism were determined (Berthelot and Bertrand 1911, 1912, Sasaki 1914, and Arai 1921).
Owing mainly to the work of Gale’s laboratory (Gale 1947) several amino acid decarboxylase preparations were described that were specific for particular amino acids. Gale studied the enzymes involved in production of amino acids by certain bacteria, their properties, and the environmental conditions required for production of these enzymes (Gale 1940). Gale also determined early applications for these enzymes including estimation of particular amino acids (Gale 1954). In 1945, Taylor and Gale made the first attempt to isolate GAD, which was followed by Umbreit and Gunsalus in 1945, and Najjar and Fisher in 1953. In 1960, Shukuya and Schwert described procedures for higher purity (90%) and identified additional properties of the enzyme (Shukuya and Schwert 1960a). Umbreit and Gunsalus were the first to propose the idea that all amino acid decarboxylases are pyridoxal phosphate (PLP) dependent (Umbreit and Gunsalus 1945). After much debate (Mardashev and Semina 1964, and Cozzani 1965), Lawson and Quinn firmly established the enzyme as being PLP dependent in 1967. Conflicting studies on GAD activity and glutamate use by E. coli led to the mapping of the GAD gene (gad) and its regulation by both closely and loosely linked genes (Halpern and Umbarger 1961, and Marcus and Halpern 1969).
In the late 1980s, the enzyme was crystallized (Markovic-Housley et al. 1987, and Sukhareva et al. 1989). In the 1990s, its amino acid sequence and genes were compared to those of other species, and it was determined GADs of various organisms probably share a common origin (Maras et al. 1992, and Smith et al. 1992). Recent research has investigated the regulation of GADa and GADb expression during acid stress (Castanié-Cornet et al. 2010) and the residues important for pH-dependent activity of the enzyme (Pennacchietti et al. 2009).
Glutamate decarboxylase is unusually specific, showing significant activity only on L-glutamic acid and α-methyl glutamic acid (Fonda 1972a). The same report shows the following compounds to be neither substrates nor inhibitors: D-glutamate, D- and L-aspartate, α-amino adipic and α-aminopimelic acids. Pure L-glutamine is not a substrate. The enzyme of E. coli shows different physical and catalytic properties from the mammalian enzyme, but has shown some similarities in inhibition studies (Youngs and Tunnicliff 1991). Inhibition studies have suggested cysteinyl residues play a role in glutamate binding to the enzyme (McCormick and Tunnicliff 2001).
GAD of E. coli exists as a hexamer of approximately 50 kDa identical subunits, each containing one molecule of pyridoxal phosphate (PLP) (To 1971).
E. coli has four inducible acid resistance systems to survive extreme acidity. GAD is part of E. coli’s second acid resistance system (AR2). This system requires extracellular glutamate and consists of the two isoforms (GADa and GADb) and a putative glutamate:g-aminobutyric acid (GABA) antiporter called GADc (Malashkevich et al. 1998, De Biase et al. 1999, Richard and Foster 2003, and Richard and Foster 2004). It is believed that this system protects the cell from acid stress by consuming extracellular protons during each decarboxylation reaction to maintain the internal pH of the cell (Castanié-Cornet et al. 1999). Richard and Foster suggested that it may be more important that the system reverses the change in transmembrane potential (Richard and Foster 2004).
Two virtually identical genes have been found (gadA and gadB), which encode the two isozymes (GADa and GADb) (Smith et al. 1992, De Biase et al. 1996, and Shul’ga et al. 1999). The protein products differ in only five amino-acid residues, and their functional properties are identical (McCormick and Tunnicliff 2001). A short length of bacterial, cat, and drosophila GAD (a 55 residue segment around the lysine responsible for covalent binding to the coenzyme) has been shown to have significant similarity (25% identity), suggesting the enzymes arose from a common origin (Maras et al. 1992). GAD from Streptococcus pneumoniae has been found to exhibit 28% homology with GAD65 from human brain (García and López 1995).
- Detection of E. coli in food and water samples (Grant et al. 2001)
- Determination of glutamate and aspartic acid (Gale 1947)
P69908
- 316 kDa (Dutyshev et al. 2005)
- 3.8 (Shukuya and Schwert 1960b; Strausbauch and Fischer 1970a)
- 5.22 (Theoretical)
- 505,620 cm-1 M-1 (Theoretical)
- E 1%, 280 = 17.0 (Strausbauch and Fischer 1970a)
- Pyridoxal phosphate is a necessary but firmly bound coenzyme. Because of the reversible configurational change at about pH 5.5 (O’Leary and Brummund 1974), chloride ions may be stimulatory to activity and acetate ions may inhibit. The crystalline enzyme has a maximum specific activity of 100-120 units/mg (pH 4.5, 38°C).
- L-isoglutamic acid, and aliphatic dicarboxylic acids, especially glutaric, pimelic and adipic acids (Fonda 1972a)
- α-(fluoromethyl)glutamic acid (Kuo and Rando 1981)
- Some sulfhydryl-group reagents: Mercuric chloride (most effective), pCMB, and DTNB (McCormick and Tunnicliff 2001)