Aspartate Aminotransferase - Manual
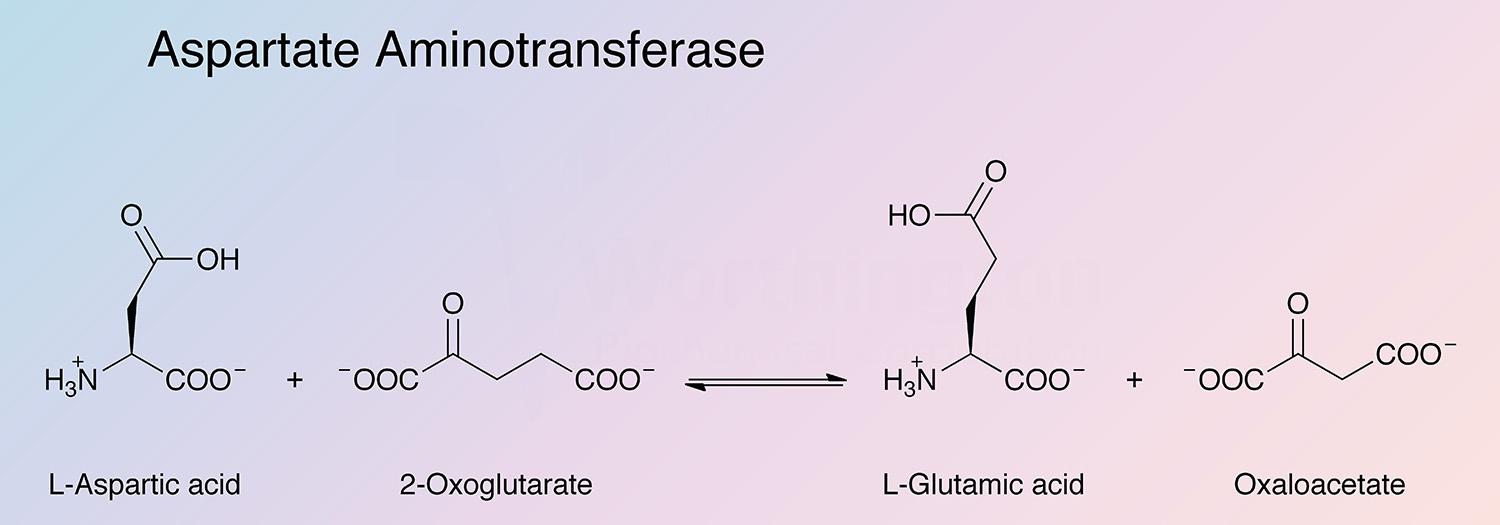
Aspartate aminotransferase (AspAT) is a key enzyme of amino acid metabolism that catalyzes the reversible transfer of the amine group from L-aspartate to 2-oxoglutarate. In mammalian tissues the enzyme is especially concentrated in heart and liver tissue. The enzyme most extensively studied is that from pig heart. The usual preparations contain only the cytosolic enzyme (cAspAT).
Transamination, the reversible transfer of the NH2 group from L-glutamate to pyruvate or other oxo acids together with an electron pair and a proton, was first demonstrated in 1937 by Braunstein and Kritzmann.
In the 1950s, the enzyme’s use in diagnostic testing for liver disease was introduced (Rej 1989). The enzyme has remained a primary diagnostic tool for liver disease for over half a century (Jeong et al. 2003).
In 1960, Fleischer et al. first reported on the separation of the two isozymes from dog heart and the differences in their substrate Kms and rate dependence on pH. In 1961, Boyd determined that the electrophoretically slow migrating enzyme was associated with the mitochondrial fraction (Martinez-Carrion et al. 1965). Research through the 1960s led to the purification of the two isozymes from a variety of tissues (Boyd 1966). In 1964, Wada and Morino purified the enzyme from porcine heart. Heart AspAT provided a unique system for studying isozyme structure and function because all forms contain a pyridoxal phosphate prosthetic group that can act as an indicator of active site events (Jenkins and D’Ari 1966b, and Martinez-Carrion and Tiemeir 1967).
The three dimensional structure of the porcine cytosolic enzyme was elucidated by Arnone et al. (Arnone et al. 1977 and 1985). In 1989, Nagashima et al. cloned and expressed porcine cAspAT in E. coli.
Recent work has investigated cAspAT’s role in fatty acid homeostasis (Tordjman et al. 2007) and protection against ischemic injury (Southwell et al. 2002). Work has also investigated the reduced activity of the enzyme as a result of crush syndrome (Desai and Desai 2008).
Aspartate aminotransferase has the highest reactivity toward L-aspartate and L-glutamate. In addition, the enzyme acts upon a number of aromatic amino acids. Non-aromatic amino acids are considerably poorer substrates (See tables in Shrawder and Martinez-Carron 1972, and Braunstein 1973, page 435).
Aspartate aminotransferase exists as two isozymes, one mitochondrial (mAspAT) and the other from the cytosol (cAspAT). Though differing markedly in primary structure, chemical and physical properties, both catalyze the same reaction with subtly different catalytic steps (Stankewicz et al. 1971, Wada et al. 1971, Watanabe and Wada 1971, and Martinez-Carrion et al. 1970).
cAspAT consists of two identical 412 residue subunits. Each subunit contains a large and small domain with the active site being located between the two domains. The pyridoxal phosphate cofactor is located in the large domain, covalently linked by an aldimine bond (Schiff base bond) to Lys258 (Rhee et al. 1997).
The gene encoding aspartyl aminotransferase (got1) is located on chromosome 14 in Sus scrofa. Nine exons encode the mature enzyme. The gene is conserved in human, chimpanzee, dog, cow, mouse, rat, chicken, zebrafish, fruit fly, mosquito, C. elegans, S. pombe, S. cerevisiae, K. lactis, E. gossypii, M. grisea, N. crassa, A. thaliana, and rice (Gene ID: 396967).
- Clinical tests for diagnosis of liver and heart disorders
- Synthesis of unnatural L-amino acids from alpha-keto acids
P00503
- Class: Alpha Beta
- Architecture: Alpha-Beta Complex; 3-Layer(aba) Sandwich
- Topology: Aspartate Aminotransferase (domain 1); Aspartate Aminotransferase (domain 2)
- Monomer: 47.0 kDa (Polyanovsky et al. 1972)
- Dimer: 93.1 kDa (Ovchinnikov et al. 1973)
- Dimer: 94.0 (Polyanovsky et al. 1972)
8.0
6.83 (Theoretical)
- 134,340 cm-1 M-1 (Theoretical)
- E1%, 280 = 14.0 (Polyanovsky et al. 1972)
- Lysine (K258)
- Pyridoxal or pyridoxamine phosphate activate undenatured enzyme (Jenkins et al. 1959)
- In solution the enzyme may be stabilized with a-ketoglutarate and EDTA in maleate or succinate buffers (Jenkins et al. 1959).
- 2-Oxoglutaconate, 2-oxoglutaconic acid dimethyl ester, and N-ethylmaleimide (Kato et al. 1996)
- N-5’-Phosphopyridoxyl L-aspartate (Izard et al. 1990)
- p-Mercuribenzoate (Stankewicz et al. 1971)
- L-a-Methylaspartic acid (Melander 1975)
- 2-Amino-3-butenoic acid (Rando 1974)
- Maleate, succinate, glutarate, and adipate (Jenkins et al. 1959)