For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
DNA Polymerase I - Manual
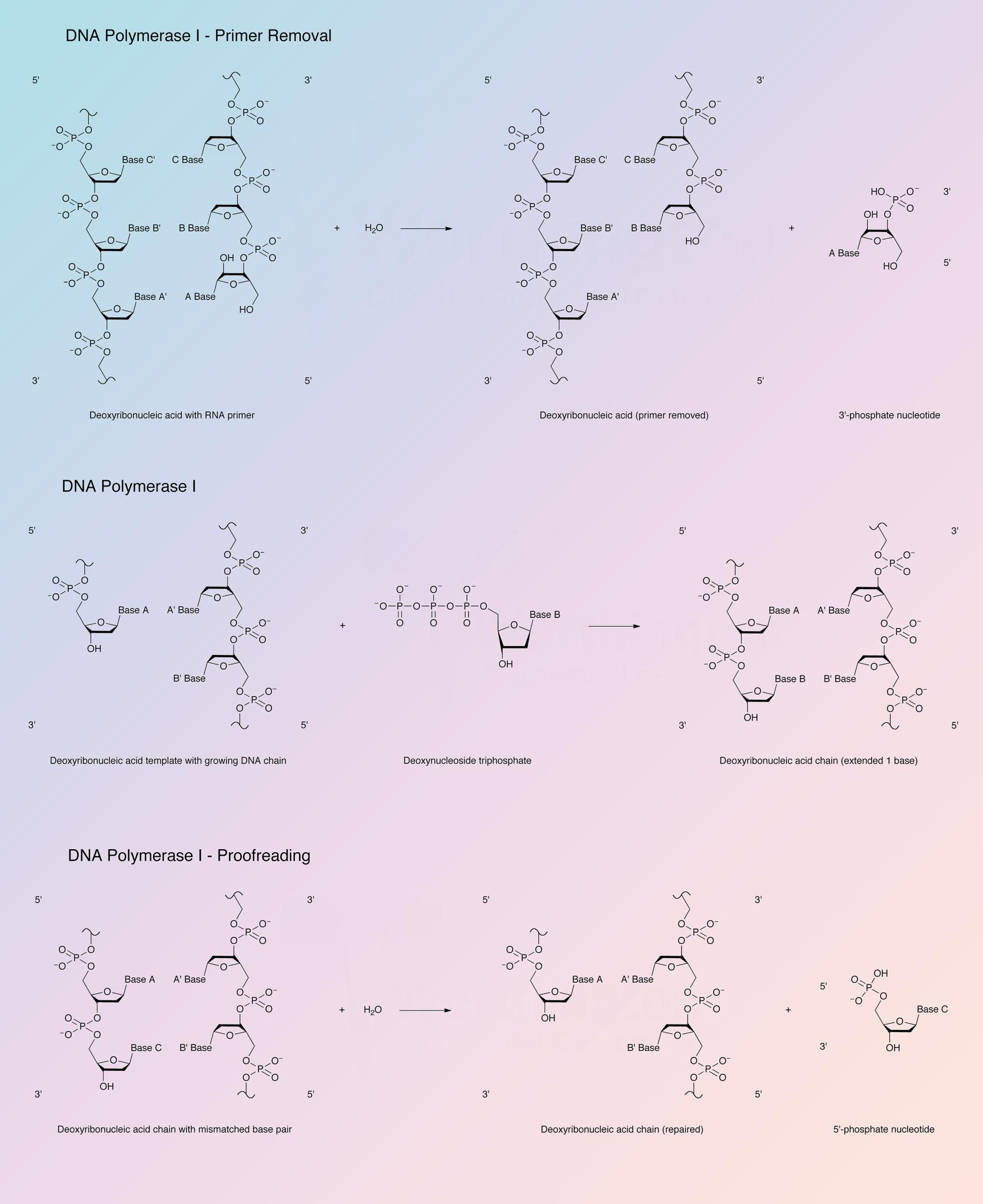
DNA polymerase I participates in the DNA replication of prokaryotes. DNA chain growth is in the 5’ to 3’ direction with addition at the 3’ hydroxyl end. The new chain is base-paired with the template, and the new chain and template are antiparallel. DNA polymerase I is the most abundant polymerase and functions to fill gaps in DNA that arise during DNA replication, repair, and recombination.
DNA polymerase I was discovered by Arthur Kornberg et al. in 1956. His initial results were first presented at the 1956 annual meeting of the Federation of American Societies for Experimental Biology (FASEB) in Atlantic City, New Jersey. Reviewers of his initial paper suggested that the authors refer to the product as ‘polydeoxyribonucleotide’ rather than ‘DNA’; ‘DNA’ was only approved after an appeal to the editor-in-chief, John Edsall (Friedberg 2006). Two more papers were published in 1958 by Lehman et al. and Bessman et al., which definitively established DNA polymerase was performing DNA replication. Kornberg was awarded the Nobel Prize in 1959 for his discovery of DNA polymerase I.
In 1969, Jovin et al. elucidated the amino acid composition (Jovin et al. 1969a, b). That same year, DeLucia and Cairns isolated an E. coli strain with a mutation that affected the DNA polymerase and surprisingly found that the mutant synthesized DNA normally. This discovery cast doubts on the role of DNA polymerase in replication and led groups to search for other replication enzymes. At the same time, Klenow and colleagues showed that the treatment of DNA polymerase with the proteolytic enzyme subtilisin (type Carlsberg) resulted in an increase of polymerase activity and decrease of exonuclease activity. The resulting DNA polymerase was isolated and was named the “Klenow fragment” (Klenow and Henningsen 1970a, and Klenow and Overgaard-Hansen 1970).
In 1970, DNA polymerase II of E. coli was isolated and characterized by Arthur Kornberg’s son, Thomas Kornberg (Kornberg and Gefter 1970). DNA polymerase II was also independently reported on by Knippers and by Moses and Richardson in 1970 (Moses and Richardson 1970b). A year later, Thomas Kornberg and Gefter identified DNA polymerase III (Kornberg and Gefter 1971).
Recent work with DNA polymerase I has included investigating the molecular basis of substrate specificity through thermodynamic studies (Wowor et al. 2010) and single-molecule FRET experiments (Santoso et al. 2010). Hastings et al. have investigated the interactions of the five E. coli DNA polymerases during cellular stress (Hastings et al. 2010), and Kukreti et al.’s studies have aimed to determine which residues are important for 3’-5’ exonuclease activity (Kukreti et al. 2008).
DNA synthesis requires a primer strand with a free 3’-hydroxyl terminus annealed to a DNA template strand and the deoxynucleotide triphosphates form base pairs with the template. Addition is in the 5’ to 3’ direction with release of pyrophosphate. The enzyme is active with DNAs containing single stranded gaps and also with DNAs with single-strand breaks or nicks. Under some conditions, RNA-DNA hybrids and an RNA duplex may serve as template-primer (Setlow 1972).
The 5’ to 3’ exonuclease activity associated with DNA polymerase I degrades both single and double stranded DNA in the 5’ to 3’ direction, yielding 5’-mononucleotides. The 5’ to 3’ exonuclease activity is specific for double stranded DNA, yielding 5’-mononucleotides and oligonucleotides. DNA polymerase I can also excise mismatched regions in DNA (Setlow 1972).
The similar structure of DNA polymerases has indicated that most DNA polymerase enzymes use an identical two metal ion-catalyzed polymerase mechanism. One metal ion activates the primer’s 3’-OH for attack on the a-phosphate of the dNTP. The other metal ion stabilizes the negative charge of the leaving oxygen and chelates the b- and g-phosphates (Steitz 1999).
The Klenow fragment is a proteolytic product of E. coli DNA polymerase I that retains polymerization and 3’ to 5’ exonuclease activity, but has lost 5’ to 3’ exonuclease activity.
DNA polymerase I is the predominant polymerizing enzyme found in E. coli. It contains a single disulfide bond and one sulfhydryl group (Jovin et al. 1969b). Five distinct DNA polymerases have been isolated from E. coli and have been designated I, II, III, IV, and V. DNA polymerase I functions to fill DNA gaps that arise during DNA replication, repair, and recombination. DNA polymerase II also functions in editing and proofreading mainly in the lagging strand (Kim et al. 1997, Wagner and Nohmi 2000). DNA polymerase III is the main replicative enzyme. DNA polymerase IV and V have large active sites that allow for more base misincorporation, and are therefore more error-prone. They also lack proofreading-exonuclease subunits to correct misincorporations (Nohmi 2006, and Hastings et al. 2010). DNA polymerase V is present at significant levels only in SOS-induced cells and over-expression restricts DNA synthesis (Marsh and Walker 1985).
The domain shape of all polymerases whose structures are known has been described as a “right hand” with “thumb”, “palm”, and “finger” domains (Kohlstaedt et al. 1992). The palm region is thought to catalyze the phosphoryl transfer, and the finger region is thought to interact with the incoming nucleoside triphosphate and the template base it is paired to. The thumb is believed to help in positioning the DNA and in translocation (Brautigam and Steitz 1998).
The gene encoding DNA polymerase I (polA) contains approximately 3,000 base pairs and encodes approximately 1,000 amino acid residues in a simple polypeptide chain. Even organisms separated by a billion years of evolution (such as Deinococcus-Thermus genera and E. coli) have approximately 35% amino acid identity and approximately 50% homology (Patel et al. 2001).
- High percentage incorporation of radioactivity for nick translation assays
- Standard reference material for the study of DNA polymerases
- Manufacturing of alternating copolymers such as poly d(A-T) and homopolymers such as poly dG-poly dC
- Klenow fragment: DNA sequencing (Sanger et al. 1977), fill-in of 5’ overhangs and removal of 3’ overhangs to form blunt ends (Sambrook 1989), and second strand synthesis in mutagenesis (Gubler 1987)
P00582
- 109 kDa (Jovin et al. 1969a, b)
- Klenow fragment: 70 kDa (gel filtration, Klenow and Overgaard-Hansen 1970)
- Maximum activity is obtained at pH 7.4 with potassium phosphate buffer for native DNA or poly dAT template-primer systems (Richardson et al. 1964)
- Klenow fragment: Maximal activities are obtained at 7.4 with phosphate buffer and at 8.4 with Tris-HCl buffer
- 5.40 (Theoretical)
- 81,030 cm-1 M-1 (Theoretical)
- E1%, 280 = 7.86 (Theoretical)
- In 10 mM sodium bicarbonate, the A280/A260 ratio is 1.81 and the absorbance at 280 nm of a 1 mg/ml solution is 0.85 (Jovin et al. 1969a)
- A divalent cation is required for activity
- Mg2+ at a concentration of 7 mM yields optimum activity under the conditions of the standard assay (Richardson et al. 1964)
- Mn2+ can partially fulfill the metal ion requirement
- Enzyme activity is also influenced by concentrations of monovalent cations such as K+, Rb+, Cs+, and NH4+
- Kanchanomycin, mitomycin, bleomycin, phleomycin, ananthramycin, plurmycin A (Tanaka et al. 1965), and neomycin (Lazarus and Kitron 1973)
- Actinomycin inhibits only when guanosine and cytosine nucleotides are present (Cohen and Yielding 1965)
- Dideoxynucleoside
- Arabinosyl nucleotide triphosphate
- Deoxyuridine-5’-triphosphate and analogues of uridine and deoxyuridine with 5’-hydroxy or amino substituents (Kornberg 1974)
- Chloroquine and some of its analogs (Cohen and Yilding 1965)