For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
DNA Polymerase, T4 - Manual
Bacteriophage T4 DNA polymerase is a DNA-directed 5’ to 3’ DNA polymerase. It is the product of gene 43 of the bacteriophage T4, and is therefore often referred to as T4 gp43 DNA Polymerase.
Bacteriophages were used by Alfred Hershey and Martha Chase in early experiments to support the theory of DNA as the genetic material (Hershey and Chase 1952, and Hershey 1953).
Throughout the 1960s, the genetic studies on T4 mutants by Epstein, Edgar and their colleagues led to the construction of a T4 phage genetic map (Epstein et al. 1963, Edgar et al. 1964, and Edgar and Lielausis 1964). Additional experiments with T4 showed that replication fidelity was determined by nucleotide selection accuracy during polymerization, and by a balance between polymerization and 3’ to 5’ excision/proofreading (Goodman et al. 1993, and Nossal 1998). In 1965, de Waard et al. demonstrated that T4 DNA polymerase was encoded by gene 43. Soon after, Speyer et al. found that mutation frequencies in unlinked genes were increased after infection with phage and some polymerase mutations (Speyer et al. 1966). These findings led to the surprising discovery that some polymerase mutations could act as “antimutators” and decrease the frequency of other mutations (Drake and Allen 1968, and Drake et al. 1969).
The replication complex of bacteriophage T4 was one of the first systems to be successfully reconstituted in vitro (Morris et al. 1975, and Young et al. 1992). Throughout the 1970s, the laboratories of Bruce Alberts and Nancy Nossal were instrumental in identifying the seven T4 coded gene products as well as the single stranded DNA binding protein (Nossal 1979, and Nossal and Peterlin 1979).
T4 DNA polymerase found its use in molecular biology because, unlike other DNA polymerases, it does not have the ability to extend from a nick, which is important in site-directed mutagenesis (Henikoff 1990). T4 DNA polymerase’s lack of 5’ to 3’ exonuclease activity also prevents digestion of annealed mutagenic oligonucleotides and reversion to parental sequence (Doetsch 1985).
Recent research on T4 DNA polymerase mutants has identified a new motif in the family of B DNA polymerases (Li et al. 2010). Nelson and Bencovic have also studied the activity of the enzyme during DNA replication after DNA lesions are introduced (Nelson and Benkovic 2010).
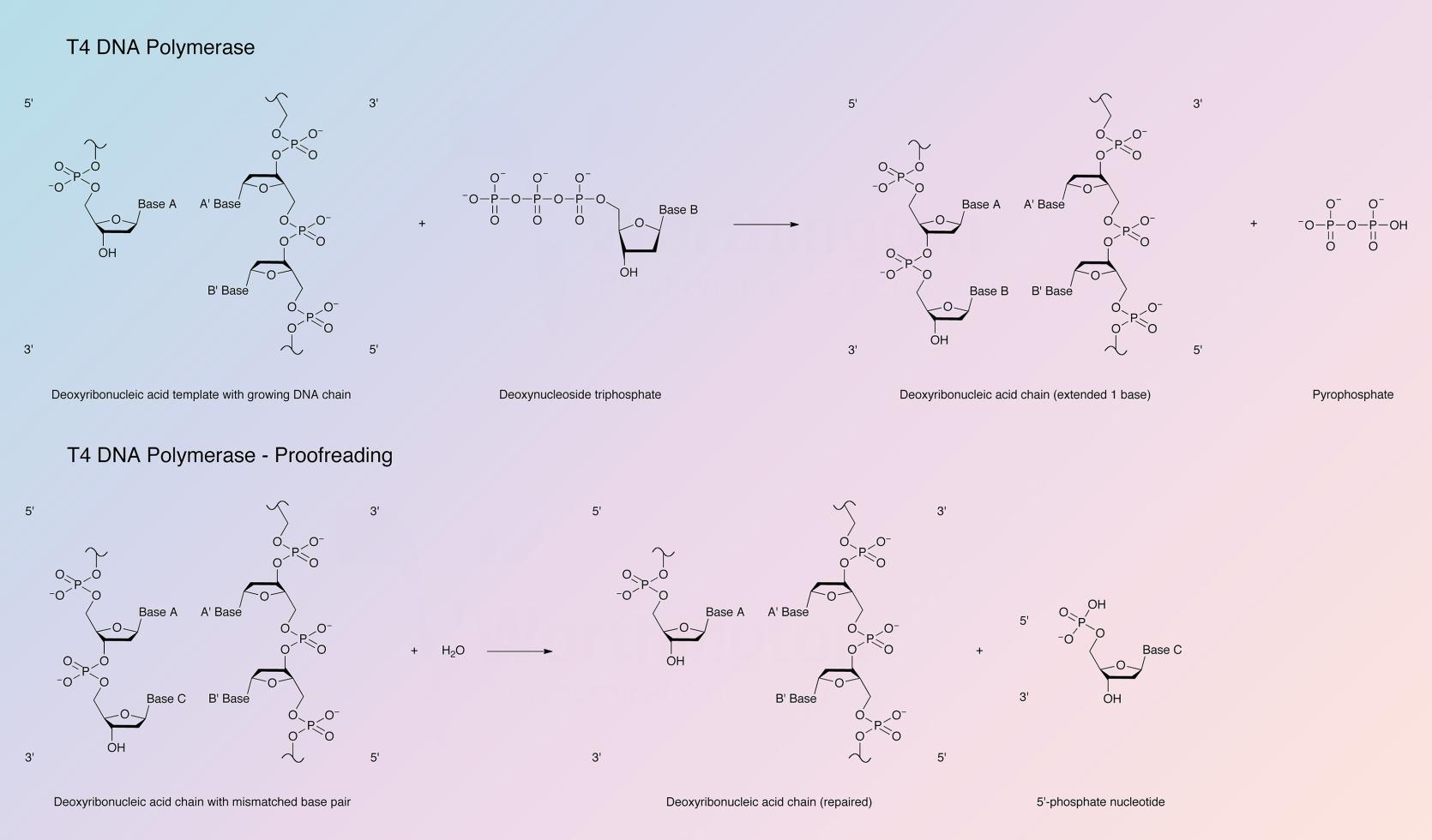
The enzyme catalyzes the polymerization of deoxynucleotide triphosphates in a 5’ to 3’ direction. It possesses very active 3’ to 5’ exonuclease activity that is more active on single than double stranded DNA; T4 DNA polymerase has no 5’ to 3’ exonuclease activity. For polymerase activity the enzyme requires DNA with a 5’ protruding end and a high concentration of dNTPs. In the presence of a high concentration of dNTPs the exonucleolytic activity is inhibited by the polymerase activity. The enzyme is not active with intact double stranded DNA as the template. For polymerase activity the enzyme requires a primed single strand of DNA, a duplex DNA with gaps, or a single stranded DNA with protruding 5’ termini.
Bacteriophage T4 DNA polymerase is also capable of an exchange (replacement) reaction. In the presence of only one dNTP the 3’ to 5’ exonuclease will degrade double stranded DNA from the 3’ hydroxyl terminus until a base is exposed that is complementary to the dNTP present. A continuous series of syntheses and exchange reactions will take place at that position.
The bacteriophage T4 encodes ten proteins known collectively as the replisome. These proteins are responsible for the replication of the phage genome and are divided into three activities: replicase, primosomal, and Okazaki repair. T4 DNA polymerase is part of the replicase, along with the gene 45 sliding clamp, the gene 44 and 62 encoded ATP-dependent clamp loader, and the gene 32 single stranded DNA binding protein (Frankllin et al. 2001, and Mueser et al. 2010). T4 DNA polymerase is active as a monomer, but it has been suggested that dimerization is necessary for coordination of leading and lagging strand synthesis (Salinas and Benkovic 2000).
The T4 DNA polymerase is an 898 amino acid residue protein. It is related to the Pol B family, which includes eukaryotic polymerases a, d, and e. The structure of RB69 DNA polymerase (a 903 residue protein) has been solved (Wang et al. 1997, and Franklin et al. 2001). Sequence alignment shows the two polymerases are 62% identical. The N-terminal domain of T4 DNA polymerase consists of residues 1-102 and 340-380. The C-terminal domain contains a PCNA interacting peptide (PIP box) motif at residues 883-903 which interacts with the sliding clamp protein (Mueser et al. 2010).
The bacteriophage T4 consists of an icosahedral head that contains the 166 kb genome and a tail segment that contains tail fibers responsible for the virus binding to the surface of E. coli.
- Filling or labeling recessed 3’ termini created by digestion of DNAs with restriction enzymes (Richardson 1964)
- Radioactive labeling of the 3’ termini of DNA (Goulian et al. 1968): The advantage is that DNA labeled by using T4 enzyme lacks the hairpin structures that can be produced during nick translation, and the labeled DNA can easily be converted into strand specific probes by cleavage with suitable restriction enzymes (Challberg and Englund 1980). The disadvantage is that this method does not produce a uniform distribution of label along the length of the DNA.
- Conversion of duplex DNA fragment ends to a double-ended structure suitable for blunt ended ligation in cloning
- Detection of stable DNA lesions (Doetsch 1985)
P04415
103.6 kDa (Spicer et al. 1988)
8.0-9.0. At pH 7.5 and pH 9.7 the activity is only 50%
- 5.93 (Theoretical)
- 130,120 cm-1 M-1
- E1%,280 = 12.56
- Asp 411, 621, 622, 684, and 686 (a cluster of aspartate residues)
- Requires Mg2+ and sulfhydryl reagents for activity
- 4-Hydroxymercuribenzoate (Lehman 1974)
- Butylphenyl nucleotides (Khan et al. 1994)
- Aphidicolin (Khan et al. 1994)
- Pyrophosphate analogs (Khan et al. 1994)
- Pyranicin (Takahashi 2008)