For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Leucine Aminopeptidase - Manual
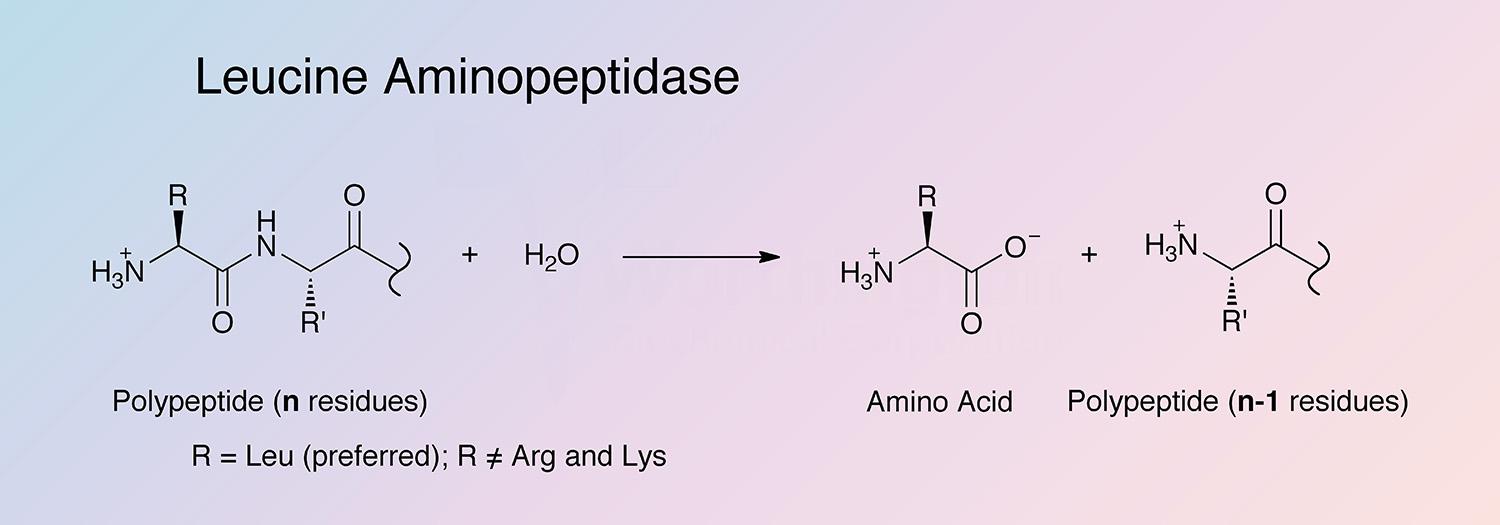
Leucine aminopeptidase (LAP) is an exopeptidase that catalyzes the hydrolysis of amino acid residues from the amino terminus of polypeptide chains. LAPs are widely distributed, ubiquitous in nature, and are of critical biological importance because of their role in protein degradation (Burley et al. 1990).
In 1929 it was reported that an enzyme found in the mucosa of pig intestine cleaved leucylglycine approximately 20 times faster than it cleaved glycylglycine. This enzyme was named dipeptidase II (Linderstrøm-Lang 1929). It was later referred to as aminoleucylpeptidase (Holter 1979, and Sträter and Lipscomb 2004).
In 1936, the enzyme was partially purified and called leucylpeptidase (Johnson et al. 1936). Johnson et al. also demonstrated that LAP is activated by magnesium and manganese.
Through the 1950s and 1960s, studies on swine kidney LAP determined chemical and physical properties of the enzyme and the zinc-metalloenzyme nature of LAP (Spackman et al. 1955, Smith and Spackman 1955, and Himmelhoch 1969).
In 1937, Sumner and Dounce first reported on the crystallization of catalase from beef liver. Along with the needles of catalase there were nearly always colorless crystals that took the shape of footballs, which lead to this protein being called football protein (FTBL protein). In unpublished work Dounce separated it from catalase. It was not until the late 1980s, fifty years after its initial discovery, that the FTBL protein was finally identified as beef liver leucine aminopeptidase (Dounce and Allen 1988).
In 1990, leucine amino peptidase became the first di-zinc enzyme for which a crystal structure at atomic resolution was determined (Burley et al. 1990).
Recent work with LAP has included determining the crystal structure of LAPs from bacterial sources (Jia et al. 2010, and Kale et al. 2010). Sus scrofa LAP has also been used as a model system for the development of molecular chaperones (Laslo et al. 2009). In addition, elevated serum levels of LAP have been found in obstructive jaundice, liver cirrhosis, liver carcinoma, and later stages of pregnancy.
The enzyme liberates amino acids from the N-terminal end of a number of proteins and polypeptides, reacting most rapidly with leucine residues (Spackman et al. 1955, and Smith and Spackman 1955). Many aliphatic amides are also hydrolyzed, as are thioesters (Metrione 1972).
Cattle eye lens LAP is a homohexamer. The monomer has a mixed alpha + beta structure. The interior of the hexamer contains six active sites that line a disk shaped cavity. Accessibility to this cavity is provided by solvent channels. This feature explains why the enzyme does not cleave longer peptides or proteins (Kim et al. 1974). Each active site contains two zinc ions, with one water ligand bridging the two ions (Burley et al. 1991, and Sträter and Lipscomb 1995).
The primary structure of porcine leucine aminopeptidase has not been determined. However, the sequence from bovine eye lens was determined by Cuypers et al. in 1982. Bovine kidney LAP cDNA studies have indicated that a 26 amino acid extension at the amino terminus is present, and not found in the mature, purified lens LAP (Wallner et al. 1993).
- Sequence analysis
- Serum control in protein studies
- Determination of L-peptides and amino acid amides containing N-terminal leucine or proline
- Cleavage of deferriform of albomycins
- Resolution of gamma-methyl and gamma-fluoroglutamic acids
(White and White 1997)
- P28839 (Sus scrofa sequence fragment)
- P00727 (Bos taurus full sequence)
326 kDa (Kretschmer et al. 1965)
9.0-9.5 (Spackman et al. 1955, and Smith and Spackman 1955)
- 6.07 (Theoretical, Bos taurus)
- 347,280 cm-1 M-1 (Theoretical, Bos taurus)
- E1%,280 = 10.28 (Theoretical, Bos taurus)
- Mg2+ or Mn2+ (3-4 mM) is essential for activity (Bryce and Rabin 1964b)
- Cd2+, Cu2+, Hg2+, and Pb2+
- EDTA
- Alcohols
- p-Chloromercuribenzoate
- Bestatin (Burley et al. 1991)
- Orthophenanthroline (Himmelhoch 1969)
- Bipyridyl (Himmelhoch 1969)
- Cupferron (Himmelhoch 1969)
- Sodium diethyldithiocarbamide (Himmelhoch 1969)
- Sodium sulfide and sodium cyanide (Himmelhoch 1969)